The goal of this document is to add a “xTAG” or a bead specific nucleotide sequence to the discovered Allele Specific Polymerase Extension (ASPE) oligonucleotides from the last blog. These are used for the detection of Single Nucleotide Polymorphisms (SNP) or mutations that leave a base-change associated with disease. There are several way to find the SNP’s that correlated to disease. It is important to remember that no all SNP’s cause disease and not all mutations that cause disease are SNP’s. This is just meant to be a basic example and use a canine model of cancer called Histocytic Sarcoma that has specific SNP’s in the PTPN11 gene that correlate to disease progression. This document will use the cDNA for simplicity for the r-markdown document and to speed up processing. This can easily be done with a genomic segment and a future blog will show the use of the OMIM-ClinVar database with the aspe discovery function in synxrna. This is an excerpt of a vignette to be included with the synxrna package.
Document summary:
Attaching xTAG sequences to an ASPE data set
Written by: Eric W. Olle
For use in single or multiplex ASPE assays. Designed for use with Luminex BBA systems.
CC-BY-SA-NC license for the ASPE and xTAG-ASPE data generated.
CC-BY-SA license for the r-markdown document.
GPLv3 for the code.
The xTAG file was obtained from Luminex and slightly modified to make filtering easier.
The MagPlex-TAG sequence file is from Luminex and used for teaching purposes only.
No Warranty Provided
Created: Jan 2022
Edited for blog: Aug. 26, 2022
The GitHub for this project contains the R-markdown and the data used to generate this blog. The data and the markdown document are CC-BY-SA. The license of the xTAG file is unknown. Contact your local Luminex representative.
Loading Packages
As with most R programs additional packages need to be loaded. This document will require a lot of the basic “Tidyverse” packages. They will be loaded individually. This example will not require synxrna but a lot of the initial data will be present in the data directory once published to CRAN. TmCalculator will be used to show the total Tm of the oligo + xTAG but is not required.
library(dplyr)
library(ggplot2)
library(knitr)
library(purrr)
library(stringr)
library(TmCalculator)
Background
This blog will be a continuation of the ASPE discovery in the canine PTPN11 gene. The goal of this is to show how oligonucleotides discovered in this blog can have a bead specific tag added to make multiplexing easier. The addition of a TAG is not required but makes multiplexing ASPE assays easier by not requiring individual oligo’s being attached to specific beads. Each application will be different and SynXBio Inc and the author of this document highly recommend that you directly contact Luminex for additional support.
Loading the ASPE data set
In the last blog how to generate ASPE oligo’s in R using the synxrna package was demonstrated. The length adjusted ASPE oligo’s were saved and will be loaded to show how to attach the bead specific tag. These are meant to be a teaching data set only, therefore, the cDNA sequence and not the genomic sequence was used. The ASPE assay is designed to be used primarily with genomic DNA. In a future blog the use of: synxrna package, mutation database with genomic DNA will be shown. This is meant to be a introduction and while cDNA could be used to determine the presence of the mutation in mRNA it is not the intended use.
ASPE_oligo dir name Tm 1 TATAATACTGGACCAACTT reverse canPTPN11_E76K 41.27132 2 ATATAATACTGGACCAACTT reverse canPTPN11_E76K 42.20290 3 CATATAATACTGGACCAACTT reverse canPTPN11_E76K 44.99814 4 CCATATAATACTGGACCAACTT reverse canPTPN11_E76K 47.53927 5 TCCATATAATACTGGACCAACTT reverse canPTPN11_E76K 48.07682 6 TTCCATATAATACTGGACCAACTT reverse canPTPN11_E76K 48.56957
The head() was done to verify the correct data set loaded. The next steps are taken directly from a prior blog and will generate a table and graph to look at the ASPE distribution by SNP.
Summary table of the ASPE set
Table: Summary of the ASPE oligonucleotides in the aspe_set data frame.
| name | n | min_Tm | max_Tm | mean_Tm | sd_Tm |
|---|---|---|---|---|---|
| canE69K_WT | 14 | 47.74501 | 60.37165 | 54.39219 | 3.896502 |
| canE76K_WT | 14 | 43.42922 | 59.09040 | 51.74416 | 4.719917 |
| canG503V_WT | 8 | 48.02790 | 58.77247 | 54.99987 | 3.950733 |
| canK91E_WT | 18 | 43.42922 | 55.76401 | 51.14649 | 3.980133 |
| canPTPN11_E69K | 14 | 45.58711 | 59.09040 | 52.74227 | 4.168961 |
| canPTPN11_E76K | 17 | 41.27132 | 59.16004 | 51.61707 | 5.643207 |
| canPTPN11_G503V | 11 | 45.46540 | 60.47983 | 54.64570 | 4.680201 |
| canPTPN11_K91E | 17 | 45.58711 | 56.81719 | 52.44656 | 3.628126 |
Graphical overview of the data set to look at oligo Tm distribution
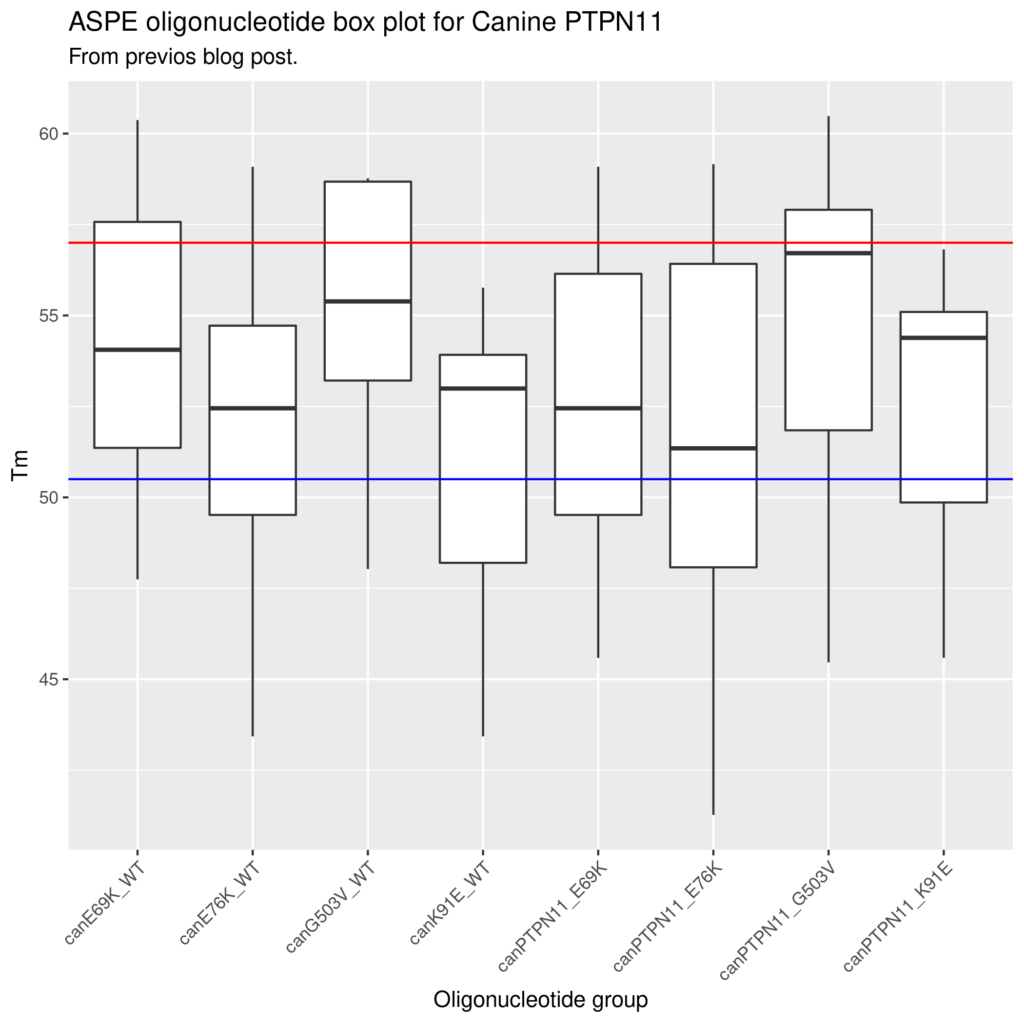
Attaching a bead specific “xTAG” for use with Luminex systems
The Luminex™ xTAG data is available from the company. This data set was copied and saved and then modified for use with R. The data used to generate this document are in the GitHub repository.
Loading the xTAG data set
This is the standard data available from the Luminex website with “instrument compatibility” first letter extracted to make selection easier. This data is for teaching purposes only and is not directly from Luminex. Please contact your Luminex representative for access to the data. The teaching data is available on the GitHub repository but is for teaching purposes only.
Reg. Part_Number Instrument_Compatibility TAG
1 1 MTAG-C001 FLEXMAP 3D Only TATTATGAGAAAGTTGAATAGTAG
2 2 MTAG-C002 FLEXMAP 3D Only TATTATGAGAAAGTTGAATAGTAG
3 3 MTAG-C003 FLEXMAP 3D Only AATAGATAAGATTGATTGTGTTTG
4 4 MTAG-C004 FLEXMAP 3D Only TGTTAAATGTATGTAGTAATTGAG
5 5 MTAG-C005 FLEXMAP 3D Only ATAGATTTAAGTGAAGAGAGTTAT
6 6 MTAG-C006 FLEXMAP 3D Only GAATGTTTGTAAATGTATAGATAG
Comp_TAG Instrument
1 CTACTATTCAACTTTCTCATAATA F
2 CTTATCATAAACTCTAACTTTCAT F
3 CAAACACAATCAATCTTATCTATT F
4 CTCAATTACTACATACATTTAACA F
5 ATAACTCTCTTCACTTAAATCTAT F
6 CTATCTATACATTTACAAACATTC F
The “eagle eyed” among the readers will notice that this has an extra column of “instrument” with a single character per row. This was done to make selection easier and there are three categories of FlexMap3d (F), Luminex200 (L) or All instruments (A).
Data filtering and xTAG attachment
The next step is to attach the xTAG sequence to the APSE Oligo’s. First the oligo’s will be filtered based upon Tm. In a “real world” application only a few oligos per mutation and WT would be chosen. This is just done to show how to generate a bunch of potentials oligo’s for use in multiplex situation.
Generating a filtered data set on Tm
#Taken from the initial ASPE oligo blog
filter_aspe_set <- aspe_set %>%
filter(Tm >= 52 & Tm < 57) #This could be narrowed to maximize similarity
knitr::kable(filter_aspe_set, caption = "Total ASPE population with Tm >=52 and <57 deg C")
Table: Total ASPE population with Tm >=52 and <57 deg C
| ASPE_oligo | dir | name | Tm |
|---|---|---|---|
| GATGTTCCATATAATACTGGACCAACTT | reverse | canPTPN11_E76K | 53.11719 |
| TGATGTTCCATATAATACTGGACCAACTT | reverse | canPTPN11_E76K | 53.35118 |
| GTGATGTTCCATATAATACTGGACCAACTT | reverse | canPTPN11_E76K | 54.93624 |
| CGTGATGTTCCATATAATACTGGACCAACTT | reverse | canPTPN11_E76K | 56.41903 |
| GTTCCATATAATACTGGACCAACTC | reverse | canE76K_WT | 52.30290 |
| TGTTCCATATAATACTGGACCAACTC | reverse | canE76K_WT | 52.59521 |
| ATGTTCCATATAATACTGGACCAACTC | reverse | canE76K_WT | 52.86586 |
| GATGTTCCATATAATACTGGACCAACTC | reverse | canE76K_WT | 54.58147 |
| TGATGTTCCATATAATACTGGACCAACTC | reverse | canE76K_WT | 54.76497 |
| GTGATGTTCCATATAATACTGGACCAACTC | reverse | canE76K_WT | 56.30290 |
| GATTACTATGACTTGTATGGAGGGA | forward | canPTPN11_E69K | 52.30290 |
| TGATTACTATGACTTGTATGGAGGGA | forward | canPTPN11_E69K | 52.59521 |
| GTGATTACTATGACTTGTATGGAGGGA | forward | canPTPN11_E69K | 54.38438 |
| GGTGATTACTATGACTTGTATGGAGGGA | forward | canPTPN11_E69K | 56.04576 |
| TGGTGATTACTATGACTTGTATGGAGGGA | forward | canPTPN11_E69K | 56.17876 |
| GATTACTATGACTTGTATGGAGGGG | forward | canE69K_WT | 53.94290 |
| TGATTACTATGACTTGTATGGAGGGG | forward | canE69K_WT | 54.17213 |
| GTGATTACTATGACTTGTATGGAGGGG | forward | canE69K_WT | 55.90290 |
| GAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 52.30290 |
| GGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 54.17213 |
| TGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 54.38438 |
| ATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 54.58147 |
| TATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 54.76497 |
| ATATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 54.93624 |
| TATATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 55.09645 |
| TTATATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 55.24665 |
| ATTATATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 55.38775 |
| TATTATATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 55.52055 |
| GTATTATATGGAACATCACGGACAATTAAAAGAGG | forward | canPTPN11_K91E | 56.81719 |
| GGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 52.59521 |
| TGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 52.86586 |
| ATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 53.11719 |
| TATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 53.35118 |
| ATATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 53.56957 |
| TATATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 53.77387 |
| TTATATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 53.96540 |
| ATTATATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 54.14533 |
| TATTATATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 54.31467 |
| GTATTATATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 55.64576 |
| AGTATTATATGGAACATCACGGACAATTAAAAGAGA | forward | canK91E_WT | 55.76401 |
| TGCGGTCTCAGAGGTCAGT | forward | canPTPN11_G503V | 52.06080 |
| GTGCGGTCTCAGAGGTCAGT | forward | canPTPN11_G503V | 54.50290 |
| GGTGCGGTCTCAGAGGTCAGT | forward | canPTPN11_G503V | 56.71243 |
| TGGTGCGGTCTCAGAGGTCAGT | forward | canPTPN11_G503V | 56.85745 |
| ATGGTGCGGTCTCAGAGGTCAGT | forward | canPTPN11_G503V | 56.98986 |
| GCGGTCTCAGAGGTCAGG | forward | canG503V_WT | 53.90290 |
| TGCGGTCTCAGAGGTCAGG | forward | canG503V_WT | 54.21869 |
| GTGCGGTCTCAGAGGTCAGG | forward | canG503V_WT | 56.55290 |
This data set could be filtered to minimize the oligo number, one selected by group or several generated and tested for activity in the lab.
Attaching the bead specific tag
In the following example, the xTAG sequence beads that will work on All (A) instruments is chosen.
filter_aspe_set %>%
bind_cols(slice_sample(filter(mag_tag, Instrument == "A" ), n = nrow(.))) %>%
cbind("tag_ASPE" = str_c(.$Comp_TAG, .$ASPE_oligo)) -> tag_aspe_set
head(tag_aspe_set)
## ASPE_oligo dir name Tm Reg. ## 1 GATGTTCCATATAATACTGGACCAACTT reverse canPTPN11_E76K 53.11719 12 ## 2 TGATGTTCCATATAATACTGGACCAACTT reverse canPTPN11_E76K 53.35118 55 ## 3 GTGATGTTCCATATAATACTGGACCAACTT reverse canPTPN11_E76K 54.93624 22 ## 4 CGTGATGTTCCATATAATACTGGACCAACTT reverse canPTPN11_E76K 56.41903 30 ## 5 GTTCCATATAATACTGGACCAACTC reverse canE76K_WT 52.30290 19 ## 6 TGTTCCATATAATACTGGACCAACTC reverse canE76K_WT 52.59521 75 ## Part_Number Instrument_Compatibility TAG ## 1 MTAG-A012 All xMAP Instruments AGTAGAAAGTTGAAATTGATTATG ## 2 MTAG-A055 All xMAP Instruments GAAGATATTGAAAGAATTTGATGT ## 3 MTAG-A022 All xMAP Instruments GATTGATATTTGAATGTTTGTTTG ## 4 MTAG-A030 All xMAP Instruments GTGTTATAGAAGTTAAATGTTAAG ## 5 MTAG-A019 All xMAP Instruments GTGTGTTATTTGTTTGTAAAGTAT ## 6 MTAG-A075 All xMAP Instruments TTTGTTAGAATGAGAAGATTTATG ## Comp_TAG Instrument ## 1 CATAATCAATTTCAACTTTCTACT A ## 2 ACATCAAATTCTTTCAATATCTTC A ## 3 CAAACAAACATTCAAATATCAATC A ## 4 CTTAACATTTAACTTCTATAACAC A ## 5 ATACTTTACAAACAAATAACACAC A ## 6 CATAAATCTTCTCATTCTAACAAA A ## tag_ASPE ## 1 CATAATCAATTTCAACTTTCTACTGATGTTCCATATAATACTGGACCAACTT ## 2 ACATCAAATTCTTTCAATATCTTCTGATGTTCCATATAATACTGGACCAACTT ## 3 CAAACAAACATTCAAATATCAATCGTGATGTTCCATATAATACTGGACCAACTT ## 4 CTTAACATTTAACTTCTATAACACCGTGATGTTCCATATAATACTGGACCAACTT ## 5 ATACTTTACAAACAAATAACACACGTTCCATATAATACTGGACCAACTC ## 6 CATAAATCTTCTCATTCTAACAAATGTTCCATATAATACTGGACCAACTC
The head() shows the basic structure of the data frame and it contains a lot of information that can be quickly exported to csv or excel. This file can then be used to order the appropriate oligo’s and beads.
Looking at a random selection of 25 xTAGed ASPE oligos
tag_aspe_set %>%
select(name, dir, Reg., tag_ASPE) %>%
slice_sample(., n = 25) %>%
arrange(name, Reg.) %>%
knitr::kable(caption = "Random sample of ASPE oligo's with Luminex specific tag sequences")
Table: Random sample of ASPE oligo’s with Luminex specific tag sequences
| name | dir | Reg. | tag_ASPE |
|---|---|---|---|
| canE76K_WT | reverse | 19 | ATACTTTACAAACAAATAACACACGTTCCATATAATACTGGACCAACTC |
| canE76K_WT | reverse | 29 | TACTACTTCTATAACTCACTTAAATGATGTTCCATATAATACTGGACCAACTC |
| canE76K_WT | reverse | 39 | ACAAATATCTAACTACTATCACAAGATGTTCCATATAATACTGGACCAACTC |
| canK91E_WT | forward | 18 | ACACTTATCTTTCAATTCAATTACTATTATATGGAACATCACGGACAATTAAAAGAGA |
| canK91E_WT | forward | 28 | CACTTAATTCATTCTAAATCTATCATATGGAACATCACGGACAATTAAAAGAGA |
| canK91E_WT | forward | 33 | ACTACTTATTCTCAAACTCTAATATTATATGGAACATCACGGACAATTAAAAGAGA |
| canK91E_WT | forward | 38 | ATTCAATACTATCTAACACTTACTTATATGGAACATCACGGACAATTAAAAGAGA |
| canK91E_WT | forward | 44 | TCATCACTTTCTTTACTTTACATTATTATATGGAACATCACGGACAATTAAAAGAGA |
| canK91E_WT | forward | 45 | TACACAATATTCATCATAACTAACGTATTATATGGAACATCACGGACAATTAAAAGAGA |
| canK91E_WT | forward | 53 | TTAACAACTTATACAAACACAAACTGGAACATCACGGACAATTAAAAGAGA |
| canPTPN11_E69K | forward | 13 | CAAATACATAATCTTACATTCACTGGTGATTACTATGACTTGTATGGAGGGA |
| canPTPN11_E69K | forward | 14 | AATTTCTTCTCTTTCTTTCACAATGTGATTACTATGACTTGTATGGAGGGA |
| canPTPN11_E69K | forward | 21 | TCAAACTCTCAATTCTTACTTAATTGATTACTATGACTTGTATGGAGGGA |
| canPTPN11_E69K | forward | 48 | AATCAACACACAATAACATTCATATGGTGATTACTATGACTTGTATGGAGGGA |
| canPTPN11_E76K | reverse | 12 | CATAATCAATTTCAACTTTCTACTGATGTTCCATATAATACTGGACCAACTT |
| canPTPN11_E76K | reverse | 22 | CAAACAAACATTCAAATATCAATCGTGATGTTCCATATAATACTGGACCAACTT |
| canPTPN11_E76K | reverse | 55 | ACATCAAATTCTTTCAATATCTTCTGATGTTCCATATAATACTGGACCAACTT |
| canPTPN11_G503V | forward | 43 | AACTTTCTCTCTCTATTCTTATTTTGCGGTCTCAGAGGTCAGT |
| canPTPN11_G503V | forward | 46 | TTAAACAATCTACTATTCAATCACGTGCGGTCTCAGAGGTCAGT |
| canPTPN11_G503V | forward | 73 | CTTTATCAAATTCTAATTCTCAACTGGTGCGGTCTCAGAGGTCAGT |
| canPTPN11_K91E | forward | 36 | ATTAAACAACTCTTAACTACACAAGTATTATATGGAACATCACGGACAATTAAAAGAGG |
| canPTPN11_K91E | forward | 52 | TTCTTCATTAACTTCTAATCTTACATGGAACATCACGGACAATTAAAAGAGG |
| canPTPN11_K91E | forward | 54 | TTAATACAATTCTCTCTTTCTCTAGGAACATCACGGACAATTAAAAGAGG |
| canPTPN11_K91E | forward | 57 | ACTTACAATAACTACTAATACTCTTATGGAACATCACGGACAATTAAAAGAGG |
| canPTPN11_K91E | forward | 63 | CTAAATCACATACTTAACAACAAATTATATGGAACATCACGGACAATTAAAAGAGG |
References
R Core Team (2022). R: A language and environment for statistical
computing. R Foundation for Statistical Computing, Vienna, Austria.
URL https://www.R-project.org/.
Wickham et al., (2019). Welcome to the tidyverse. Journal of Open
Source Software, 4(43), 1686, https://doi.org/10.21105/joss.01686
Li J (2022). TmCalculator: Melting Temperature of Nucleic Acid
Sequences. R package version 1.0.3,
https://CRAN.R-project.org/package=TmCalculator.
Olle E, W., (In Progress) The synxrna package: A tool for RNA